a Universität Oldenburg, 26129 Oldenburg; kaupp@kaupp.chemie.uni-oldenburg.de, haak@kaupp.chemie.uni-oldenburg.de
b Universität Tübingen, 72076 Tübingen; guenter.gauglitz@ipc.uni-tuebingen.de
c Universität des Saarlandes, 66041 Saarbrücken; ch12hs@rz.uni-sb.de
d CASAF Computerchemie GmbH Bitterfeld, 04425 Taucha; Dr.Rainer.Moll@t-online.de
e Schmelz, Technologie-Dienstleistungen 'ThS-tech', 72076 Tübingen; Schmelz@thstech.com
f Lab Control Scientific Consulting and Software Development GmbH, 50858 Köln; tf@labcontrol.com
Abstract:
Present habits of publication (print and electronic) are unsuitable in experimental sciences if they restrict themselves to fulltext with links and pixel graphics, because all primary data are lost. Primary high-tech data are highly interactive at the site of data production. The tools for their full communication for critical analyses, use and data mining are available and so are the data storage capacities, but these features are not yet used in publications, even though the feasibility is amply shown in databases and in the internet. It is tried to change this disastrous situation in a comprehensive way by pointing to the available data-interactive publishing devices. This covers fully interactive hypermolecules, hyperspectra, chemometric data sets, and complex 3D-objects. The data formats are XML/CML and the widespread PDB/VRML/JCAMP-DX standards rather than TIFF/GIF/JPEG projections.
The present hesitancy of authors and publishers to accept the superior techniques has to be overcome by teaching, advertizing and various helps, for the sake of science.
Keywords: data-interactive electronic publishing; primary data; molecular modelling; spectra; chemometrics; future data mining; 3D-objects; teaching; acceptance; guidance pages
1. Introduction
Common (electronic) publication techniques lead to severe losses of primary scientific data. Such uneconomic losses are no longer tolerable in view of modern possibilities of publishing and of the public sponsoring of science with the money of tax-paying people. However, many publishers and in particular authors shrink from applying new and appropriate technologies. Therefore six German research groups from academia, industry and an international publisher have set-up a comprehensive program for the development of data-interactive publishing in chemistry including natural sciences which is publicly supported by the BMBF.1
The availability of mass storage devices and innumerable communication tools in the internet and tools for interaction at the sites of the data producing instruments or in huge databases makes this endeavor timely. We thus present an outline of a new age of publication without loss of data in chemistry and natural sciences with the aim to convince publishers, authors, readers, and librarians to use it and to apply it for all sciences. The key feature is the hyperdocument that interacts with the primary data. All fields of science will profit from full disclosure of scientific data.
2. Interactive handling of molecular structures, molecular modelling
Chemists use various ways to describe their materials. They write and publish names, determine total formula and draw structural formula for print that may be converted into connectivity tables with proper software. The latter possibility, though still non-interactive, offers some advanced uses :
Table 1. Experimental atomic coordinates for o-aminophenol including the crystallographic data for the modelling of both the single molecule or the crystal packing with proprietary software (format of SCHAKAL).
| data file | chemical structure | title o-aminophenol, Acta Cryst B1979, 1394 Pbca cell 7.256 7.849 19.754 90 90 90 atom o1 -.0681 .2347 .4967 atom n1 .1941 .0835 .4213 atom c1 -.0497 .2908 .4315 atom c2 .0793 .2078 .3907 atom c3 .1067 .2589 .3245 atom c4 .0006 .3943 .2978 atom c5 -.1293 .4747 .3384 atom c6 -.1547 .4216 .4046 atom h1 .208 .194 .296 atom h2 .017 .433 .247 atom h3 -.211 .587 .316 atom h4 -.261 .491 .434 spgr P b c a pa 0 2.5 0 2 .25 .75 end |
|---|

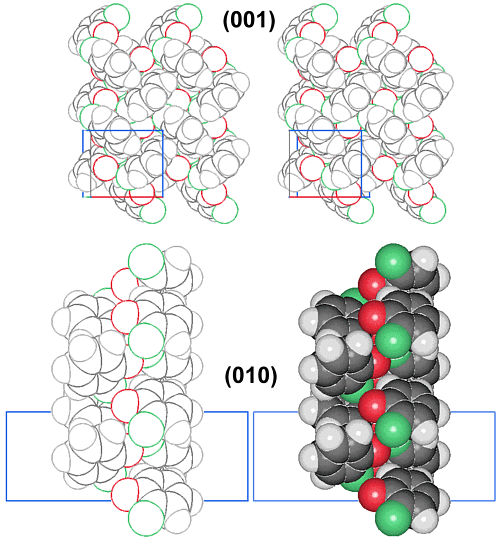
A further step is the interactive analysis of PDB format files with free software that is available in the internet, as shown in Figure 2 for the packing diagram of benzimidazole (data from CCD) on the (001) face.
 The following image is interactive, if you have the CHIME-Plugin installed ! In this case you can: Click left mouse button and drag for rotating the crystal packing or click right mouse button for more options
|
|---|
The top image is a dead TIFF, GIF, JPEG, or printed image in stereoscopic representation. However, the bottom image is interactively analyzable with mouse click if the internet server is configured for chemical-mimetypes (numerous functions in a menu: rotating, size, mono, various representations, etc.). Unfortunately, even purely electronic journals rarely use these PDB files presently, however, numerous examples can be found in internet homepages and in demos of an internet journal.2 More publishers will have to be convinced that they should offer such possibilities in their electronic publications. Furthermore, it will be essential to provide the original data (cf. Table 1) for the specialists with their proprietary software, so that full control on the structural data will be achieved also for huge protein molecules, drug design, etc.
3. Spectra
Every operator has extensive software at hand when he/she records spectra at any high-tech spectrometer and thereby interacts with the data. However, for the common (electronic) publication of these spectra, one finds oneself forced to prepare a peak list or a dead 2D-graphic with loss of all primary data which would be needed for independent analysis or for data mining. Database producers cannot use the full potential of the published spectra or even have to repeat them (including synthesis of the compounds or isolation from natural sources ). Thus, publicly sponsored important scientific data are not disclosed by common (electronic) publishing habits.
An effort has been undertaken to change that disastrous situation by data base producers with a software package TranSpec,3 which waits for being implemented to chemical markup language (CML), a subset of XML for general use. Table 2 shows the present state of the art and the aims for further development.
Table 2. TranSpec: Software for the Coverage and Treatment of Spectra and Structures
| Present State: | Aims: |
|---|---|
|
Data Coverage Quality Check Visualisation Archiving Import from external Sources Export into external Data Bases For 13C NMR H NMR IR RAMAN UV MS including structural information |
Extension to Hetero NMR ESR ORD CD etc. Inclusion of non-spectroscopic data formats, e.g. Cyclovoltammetry etc. Development of the necessary Data Exchange Formats CML-based author tools for interactive dynamic publishing
Internet-Client |
All major types of spectra are already covered, further ones and non-spectroscopic formats planned. The flow scheme of Figure 3 indicates how the spectra will be implemented into publications without loss of interactivity, i.e. without loss of data.
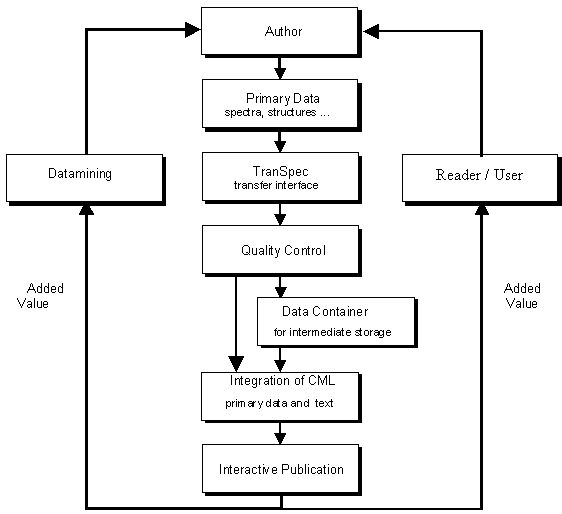
Figure 3. Flow scheme for the interactive publishing of primary data.
Authors provide their full data sets. These will undergo quality control via transfer interface and will go through storage media into interactive CML publications. This procedure provides added value for authors, readers and users. The authors will obtain a submission mask with all necessary authoring tools. They will be required to connect the structural formula with all of the primary spectroscopic data from their spectrometer. Such published spectra will be graphically searchable (via various embedded tables). They will be analyzable with mouse click and interpreted by linking to huge spectral databases or knowledge-based interpretation software. An example is given in Figure 4. Peak positions and intensities are indicated by placing the cursor. Zooming is possible, and numerous visualizations can be performed in a menu guided way, including the downloading of the file. Similar possibilities are available for different spectroscopic techniques, but not yet in publications.
|
Previous informations in publications (peaklist): FT-IR (KBr): 3580, 3090-3010, 3000-2840, 1490, 1435, 1231, 1176, 1020, 855, 753, 698 Interactive JCAMP-data:
The following image is interactive, if you have the JCAMP-Plugin installed !
|
|---|
Figure 4. An interactive IR spectrum indicating some possibilities of detailed analyses.
4. Chemometrics
Chemometric data shall be treated according to the flow scheme in Figure 3. That endeavor will require extensions of the CML-syntax in cooperation with OMF (Open Molecular Foundation). A typical scheme is shown in Figure 5.4 An array of sensors is used to generate a large set of multidimensional data (time, signal, sensor #, mole fraction) that cannot be fully represented, of course. Performance data for various sensors have to be evaluated by chemometric techniques. However, it is essential, that the raw data be preserved for future re-evaluation in different, new, more advanced ways in order to obtain better results from the data, or to extract unexpected results by data mining. Thus, such data sets in an ever increasing field shall be converted to JCAMP, HDF, NetCDF, CML and published together with the publication of the present performance values.
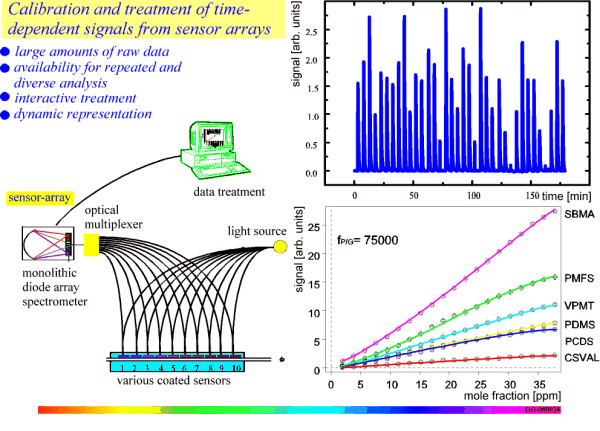
Figure 5: Creation and interactive treatment of chemometric data.
5. 3D-Objects
The interactive publication of complex 3D-data is an important topic in science and technology. Again, full data interaction is available at the high-tech sites of data generation. However, all of that is lost in common publications requiring dead pixel graphics. Thus, it is essential for a full analysis or validity checks or revised interpretations that GIF, TIFF, JPEG images be accompanied by the primary 3D-data. This will allow the user of comparable though incompatible software in different parts of the world to load the published 3D-data and view and analyze them. Thus, the XYZ and META informations in the proprietary data files have to be separated and made available for various viewers upon publication (local, VRML, 3D-CML still in preparation). A flow scheme is given in Figure 6.
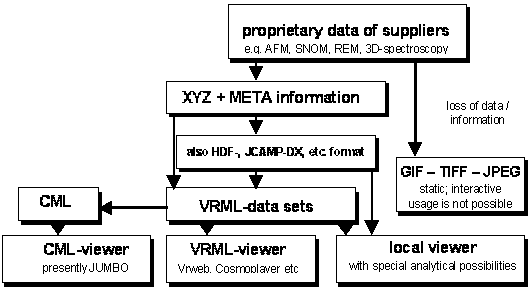
Figure 6. Flow scheme for the disclosure of 3D-data to attain interactivity.
The importance of such endeavor can be shown with an atomic force microscopic (AFM) surface. A 2D-GIF image like Figure 7 is not analyzable and does not even give a final impression of the shape of that surface. Nevertheless, thousands of images of that type reside in the published literature and do not disclose the largest part of their content.
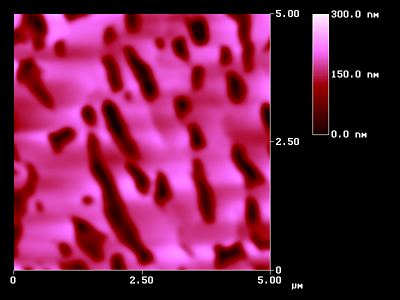
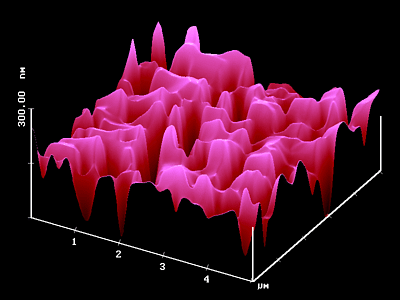
VRML Image of Fig 8 (low resolution- approx 337 kB)
VRML Image of Fig 8 (high resolution - approx 547 kB)
Table 3. Summary of important analytical and statistical modes for 3D-data to be made available in VRML, HDF, JCAMP-DX, or CML.
| Image Analysis | Statistics |
|---|---|
|
Distances Angles Heights Zoom Profile Cuts/Steepness Image Rotation Contours 3D-Images Inversions Heights |
Section Bearing Roughness Autocovariance Grain Size Depth Fractal Spectral Density Numerous Filter Functions |
It must be achieved that the 3D-data of American, European, Japanese, etc. instruments become accessible for the same interactive analysis everywhere. These and similar tasks are extremely widespread and touch all fields of science and technology. The interior of 3D-objects is disclosed by tomographic data. These are particularly well known in medical diagnostics. But even there, the DICOM exchange format is at best used in internal networks whereas full interaction with remote sites would be desirable. Of course, some tomographic databases and CD-ROM teaching products exist in medicine.
6. Acceptance by Scientists, Teaching
Whereas little problems of acceptance exist in industry, where the data are collected in local databases for retrieval and data mining, or where the wealth of publishing interactive handbooks will be another cost saving factor, the acceptance of authors, referees, readers and their publishers in academia increases but slowly. Thus, the participation of industrial partners is very important and we will not succeed if we do not instruct, teach, advertise and give all possible help to facilitate the use of the interactive electronic publishing, even though the high added value should be evident to everybody. Apart from lectures, seminars, workshops, we have to give as much help as possible in the form of interactive guidance pages that provide all of the necessary tools and easy to use software to the authors and referees. The readers gain increasing experience by their common internet surfing. An interactive guidance page lists all available types of electronic material (e.g. alphanumeric, 2D-formula, 3D-formula, 3D-coordinates, 3D-images, spectra, etc.) and the various data formats that do exist for them. The authors may convert their data into more appropriate representations with the hyperlinked tools. For example they will be guided to some free or inexpensive molecular modelling programs (e.g. AIMPAC, DeFT, Mopac, PC Gamess, Tinker, ACD Sketch etc.). In that case they may start with handwritten structural formula and have the calculation run to generate PDB-files for full animation as shown in Figure 9 for a ß-cyclodextrin complex. The pulldown menu indicates the types of interaction that may be performed. Most importantly, the interaction with a wealth of PDB data includes the downloading of the atomic coordinate table for use with proprietary programs that might have highly specialized features.
3D Animation:
Complex of b
-cyclodextrin and ANS (anilino-naphthalin-sulfonic acid)
Minimisation by CHARMm force field, charge distribution by Gasteiger-method
7. Conclusion
It will be evident that the data of the authors increase in value for the authors themselves and for the scientific community, if data interactive publication is made easy and if publishers offer that possibility. In most cases authors will certainly be prepared or can be asked to disclose all their data. Science can no longer afford to lose most of the data just by publishing certain views of the results and impeding all future data mining. Society has the right to expect that data which are produced with public support are made available for critical testing and for future use in technology, medicine and other fields.
Footnotes and References
1. This project is funded as part of the GLOBAL INFO project of the Federal Ministry of Education and Research (BMBF).
http://www.global-info.org
2. Internet Photochemistry & Photobiology 1998;
http://www.photobiology.com/IUPAC98/gkaupp/snom.html
3. TranSpec-New Strategies for Treatment of Spectra, Nachr. Chem. Tech. Lab. 1998, 46, Supplement, A 69.
4. J. Seemann, F.-R. Rapp, A. Zell, G. Gauglitz, Fresenius' J. Anal. Chem. (1997) 359, 100;
G. Kraus, G. Gauglitz, Chemometrics Int. Lab. Systems (1995) 30, 211.